Are there any "current" tools for visualizing phased haplotypes in vcf format? The whole-genome data is phased algorithmically through beagle/shapeit. It is not long read data.
The most promising one I found was inPHAP. Any other suggestions would be appreciated.


what 'type' of visualization do you want to see? if you have a screenshot of something you would like, it might guide answers
https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-15-200/figures/1
Similar to this. Essentially being able to compare loci between haplotypes.
one option: you might just want to loading the VCF data into R and plotting it as a heatmap with e.g. pheatmap! to my eye, there is not anything particularly special about the plot you linked compared to a normal heatmap, so using a R heatmap might actually be a good option :) parsing VCF is a bit tricky, but it might not be too bad
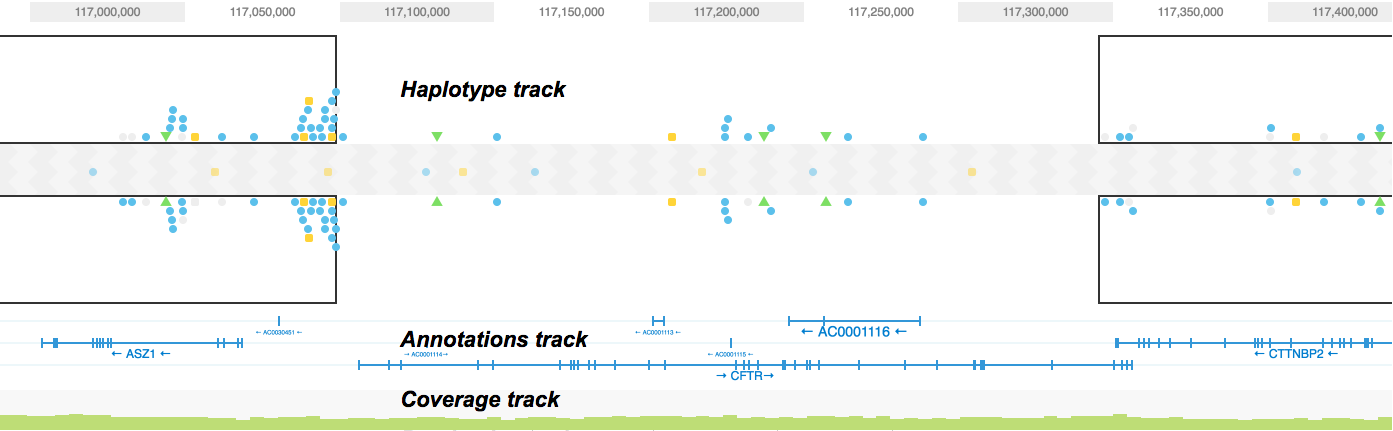
other non-R option... https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6718772/ another non-R option that I wrote, a plugin for jbrowse https://github.com/elsiklab/multivariantviewer finally, can browse this list i compile to give lots of visualization options https://cmdcolin.github.io/awesome-genome-visualization/?latest=true
note that my tool plots just 0|0 as grey (ref) 0|1 (light blue, heterozygous) 1|1 (blue, homozygous). igv also has similar types of displays, but doesn't show it as a dense matrix afaik