I'm trying to collect (and then download) all transcriptomes and genomes associated with txid7604.
The query I am using is:
((txid7604[Organism:exp]) AND ( "tsa master"[Properties] OR "wgs master"[Properties] ))
And the database I am looking at is nuccore.
When I search via the web interface I get four matches:
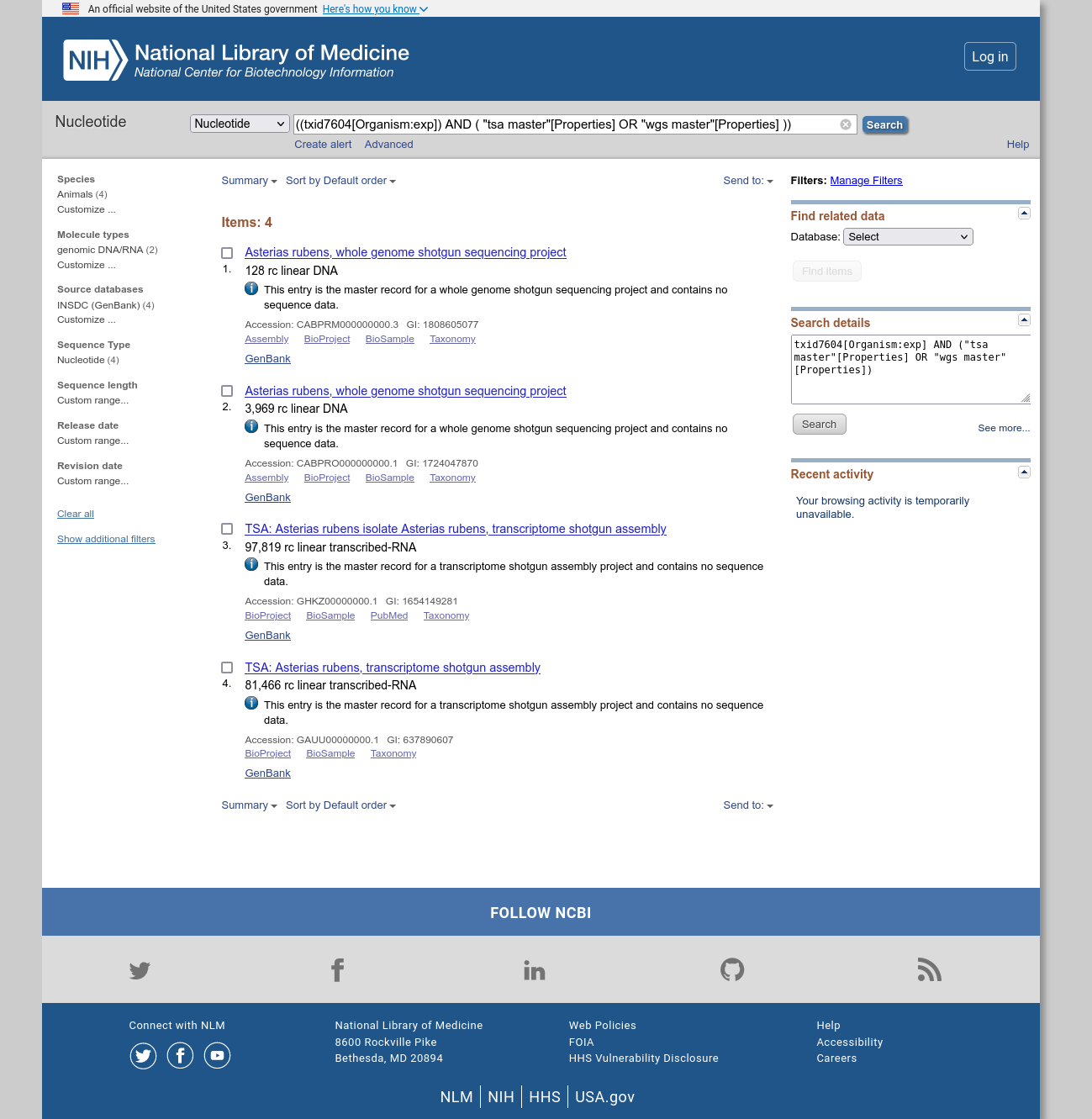
However, when I query via esearch 16.2 (installed via bioconda), I get 81468 matches?
> esearch -db nuccore -query "((txid7604[Organism:exp]) AND ("tsa master"[Properties] OR "wgs master"[Properties]))"
<ENTREZ_DIRECT>
<Db>nuccore</Db>
<WebEnv>MCID_638e1b0b374c2370e333fddb</WebEnv>
<QueryKey>1</QueryKey>
<Count>81468</Count>
<Step>1</Step>
</ENTREZ_DIRECT>
What is the source of this discrepancy?


I see. I actually managed to find out how to circumvent this.
esearch(and I suppose all its siblings) make a distinction betweentsa-masterandtsa masterandwgs-masterandwgs masterwith double quotes. Adding the hyphen resolves the discrepancy.No hyphenation -> incorrect count:
With hyphenation, correct result: