Full disclosure - I work for a company called Basepair that is a for profit bioinformatics platform provider.
It's founder came from academia where he found he was spending most of his time performing routine analyses and repeatedly answering the same questions related to NGS data analysis & visualization. He set out to create a point & click way of enabling those who were new to NGS or who had less computational experience to analyze, interact and visually explore NGS data on their own with industry standard public domain tools, setting them up to collaborate with a bioinformatician once they had a more informed question. We believe there is a place for this kind of bioinformatics solution, but would be curious as to what others think in this forum?
Anyhow, if anyone would like to try out the platform before providing feedback you can simply sign up for a free six sample trial on our website and let us know what you think. If you want to go on and use it from there there is no upfront license fee - just a pay as you go per sample usage model that includes unlimited analyses per sample and 12 months storage. What's more, if you quote 'Biostars' we'll offer between a 20% and 85% discount depending on the sample type.


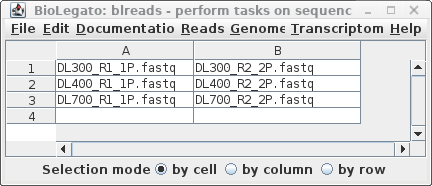
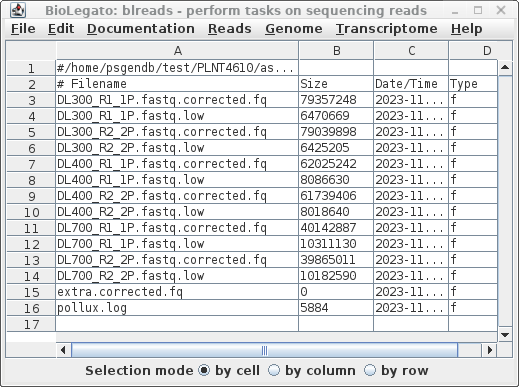

Is there any reason why your organization cannot offer a locally installable solution?
I'm not signing up on your website just to find out what is happening in the backend, but from the screenshots and blog posts (e.g., https://www.basepairtech.com/knowledge-center/a-deep-dive-into-differential-expression/) it appears you've done little more than wrap a GUI (
R-based?) around open source tools funded by public (and in some cases, personal?) funding. In this case, what portion of your profits are you using to support these open source tools and to support the development of new tooling in the future?It is also so tiring and uninspiring to have anything and everything in silico go to the "cloud" with for-profit entities subsequently trying to extract every last unit of profit by monetizing every possible aspect of the users' interactions with the software and data.
As stated in the post this is a for profit company. Supporting a locally installable solution (which may need to take into account diverse infrastructure) is likely beyond what the company can do with the resources they have (my speculation). By controlling the backend infrastructure they are able to offer a uniform user experience. Providing tech support is likely significantly more expensive when they have no control on end user infrastructure.
When one has the knowledge and resources to do something it is easy to overlook others who don't have these resources (human or otherwise) available. For them this kind of solution is the next best thing.
Thanks, this is indeed one of the reasons it is challenging to provide something that is reasonably affordable if the offering is predicated on a locally installable solution. This being said however, we do offer the option to plug in an institution's own cloud account into the platform rather than needing to upload any data to ours. I realize it's not the same thing, but it does at least mean that you remain connected to other tools and resources you have there and that data never has to leave your own IT approved environment. Not to mention of course that you can then benefit from any pre-negotiated cloud pricing your institution might have put in place.