Dear all,
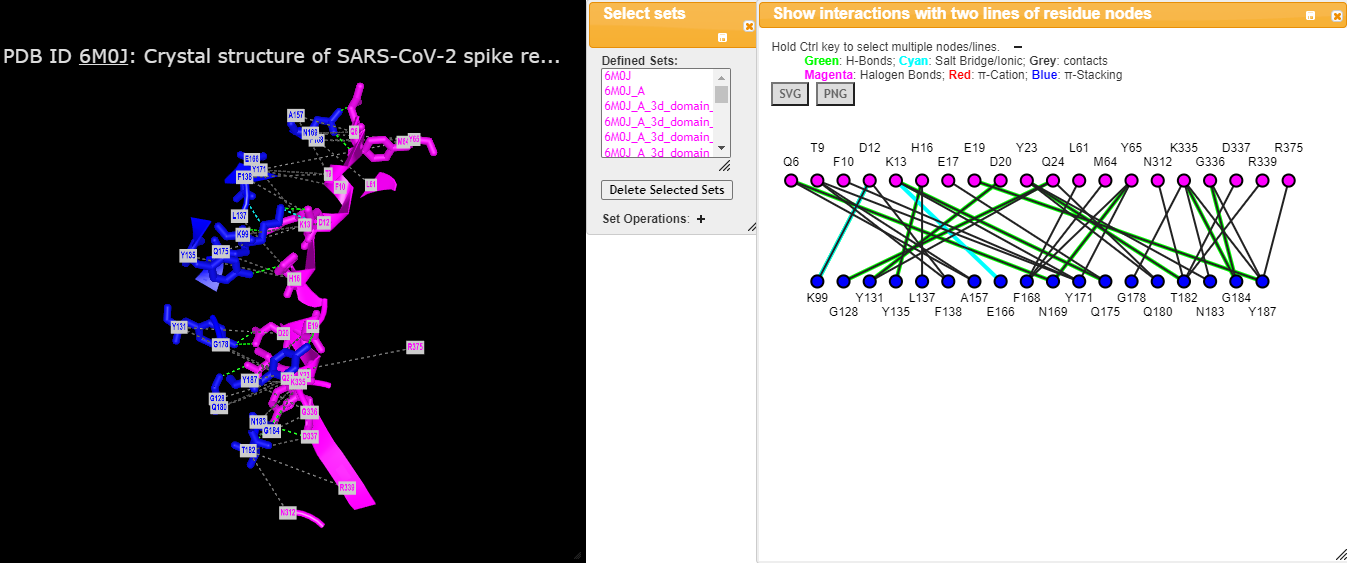
What are the best ways to study aromatic-aromatic interactions and amino-aromatic interactions between protein binding site residues and ligands in the PDB. What are the best free and paid software I can use to do an excellent job.
Thank you,
Dr. Abubakar Abdullahi.