Entering edit mode
23 months ago
cthangav
▴
110
Hello, I have a list of enhancers and the genes they target that looks like this for mm10 genome:
Enhancer Target Gene
1: chr4_98727908_98728497 L1td1
2: chr9_78623220_78624062 Cd109
3: chr9_78615332_78616035 Cd109
4: chr2_146299937_146300524 Insm1
5: chr2_146218029_146218779 Insm1
Using this list, I would like to make a plot like this, connecting each enhancer to its target gene(s):
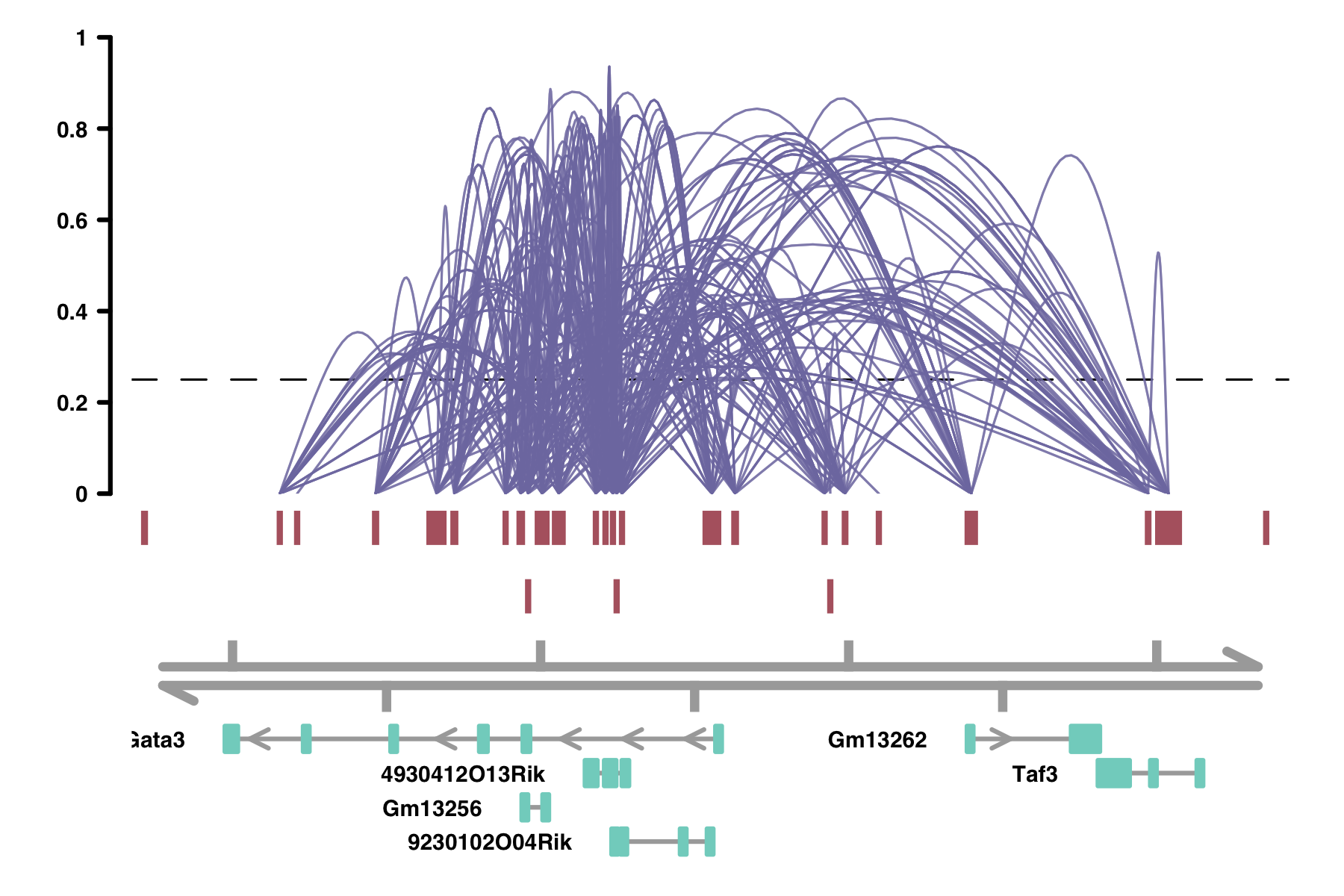
It looks like this is the function they used to make this graph: github.com/cole-trapnell-lab/cicero-release/blob/master/R/plotting.R
Is there a quick way to do this with an R package or IGV? I am not working with single cell data like Cicero is designed for.

