Entering edit mode
12 months ago
lsp03yjh
▴
860
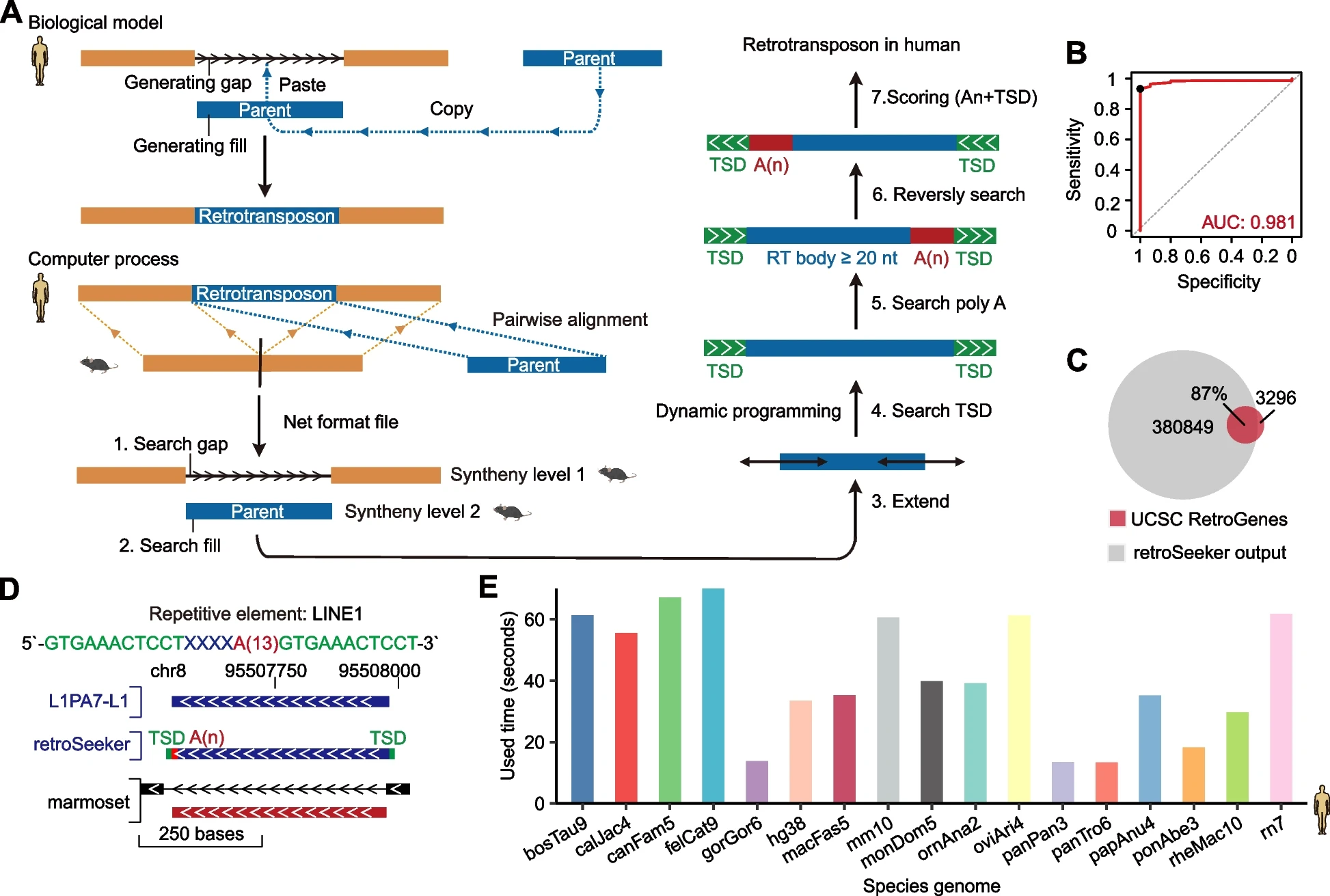
We developed retroSeeker , a new computational software that identifies novel retrotransposons from pairwise alignments of genomes and decodes their biogenesis, expression, evolution and potential functions in humans and other species.
- We discovered that the majority of new retrotransposons exhibit a specific L1 endonuclease cleavage motif, with some motifs precisely located ten nucleotides upstream of the insertion site.
- We identified that a large number of candidate functional genes might be generated through a retrotransposition mechanism.
- Importantly, we uncovered previously uncharacterized classes of retrotransposons related to histone genes, mitochondrial genes and vault RNAs.
- Moreover, we elucidated the tissue-specific expression of retrotransposons and demonstrated their ubiquitous expression in various cancer types.
- We also revealed the complex evolutionary patterns of retrotransposons and identified numerous species-specific retrotransposition events.
Taken together, our findings establish a paradigm for discovering novel classes of retrotransposons and elucidating their new characteristics in any species.
Reference
New computational approach for the discovery of retrotransposons


The link throws a 404 error. You may want to edit the post and delete the period at the end of GitHub link. Also, explaining a bit better in the repository how to create .net files would be helpful.
Thank you very much for your comments. I have corrected the GitHub link.