Entering edit mode
12 months ago
andre.arrudalima
▴
60
Hi,
I'm trying to do a protein sequence aligment of orthologues of a ~1000 aminoacid (aa) protein, in R. Normal MSA tools display boxes for each aa, which makes the aligment figure too big for my interests. The only MSA tool that I found that display aa as lines is this ggtree, but I dont need the tree.
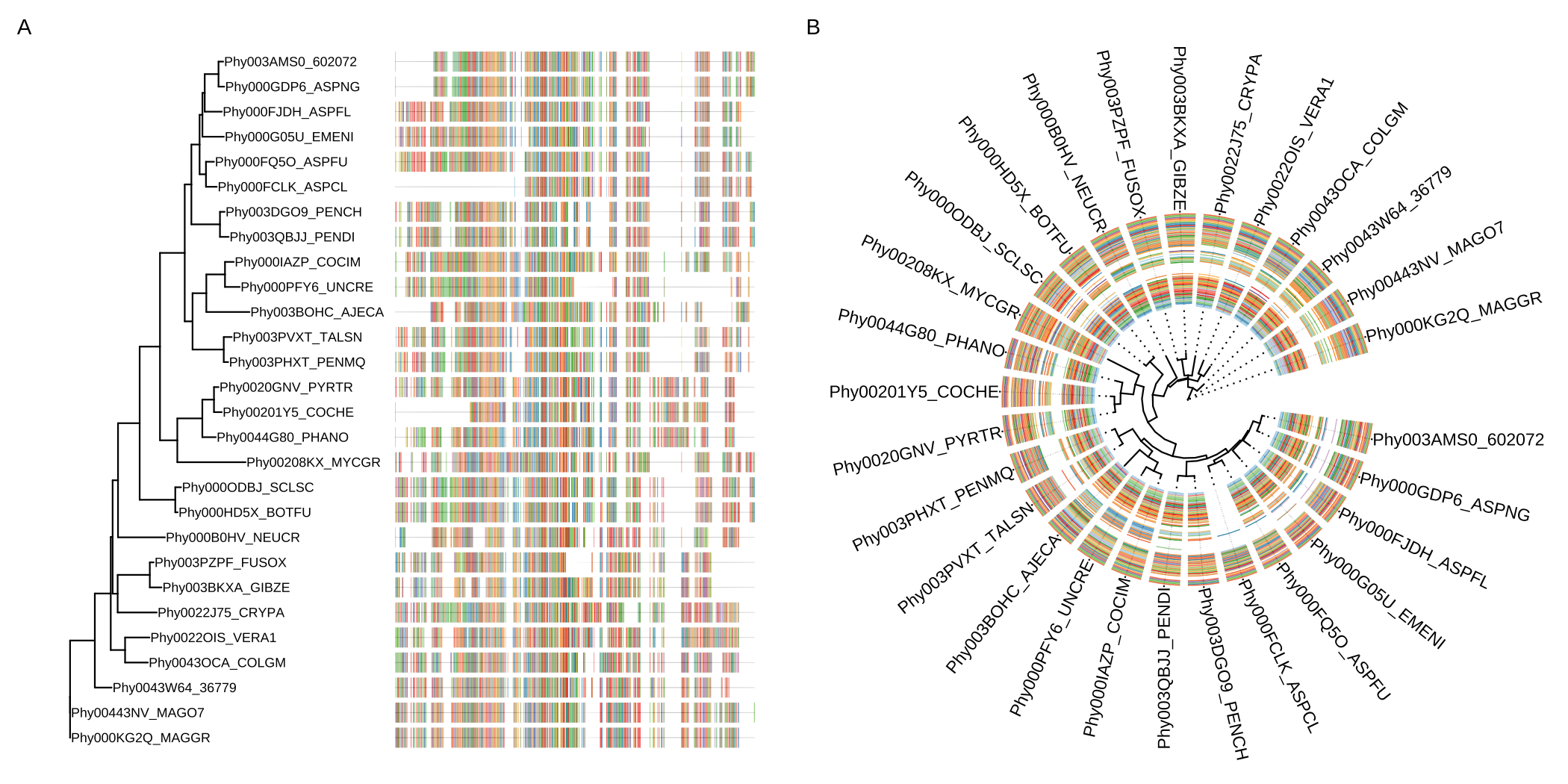
I'm having real trouble to follow the steps present here https://yulab-smu.top/treedata-book/chapter7.html and, in a more simple way, here https://rdrr.io/bioc/ggtree/man/msaplot.html#google_vignette.
So I have some questions:
- Is there a simple way to do a MSA figure that restrict each aa size? Maybe a different program.
- And how can I impor my aligment file and tree file to R?
Thank you


https://yulab-smu.top/ggmsa/