Hi all,
I recently downloaded HMMER to use hmmscan locally in Linux with a Pfam database. It works great, however the ouput files are quite difficult to read quickly in my opinion...
I tried using output options such as: --tblout, --domtblout, --pfamtblout, etc. but the ouput files are still voluminous.
I would like to keep only the top hits in my output files.
I've seen that it was possible with hmmsearch so I was wondering if there was something similar with hmmscan...
Ideally, I would want an output as I could find online: 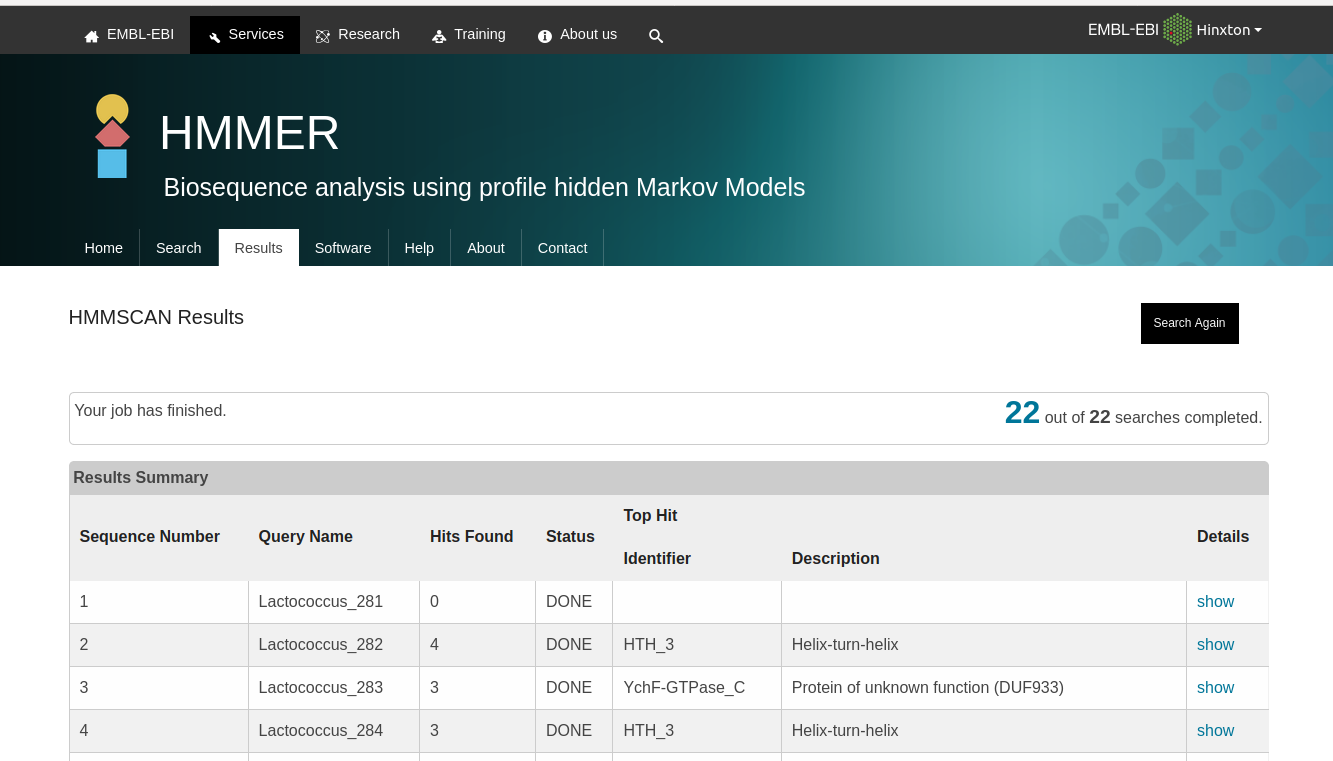
If you have any suggestions, I'll gladly took them. Thank you in advance for your very appreciated help!


Hey! You mentioned you know how to find the top hits from hmmscan. Could you share how you do this?
I'll leave a clarification. Since the link doesn't work.
Because hmmscan --tblout generates a table with the group of hits at the top with the best score, awk just leaves the first top one.