Entering edit mode
11.4 years ago
umer.zeeshan.ijaz
★
1.8k
Yesterday, I wrote new perl one-liners to emulate the functionality of FastQC. For visualisation, I am using gplot, a wrapper around gnuplot, that facilitates us to pipe quantitative data directly into gnuplot without the need of generating temporary files. You can find these utilities here (I have wrapped them in shell scripts for ease of use):
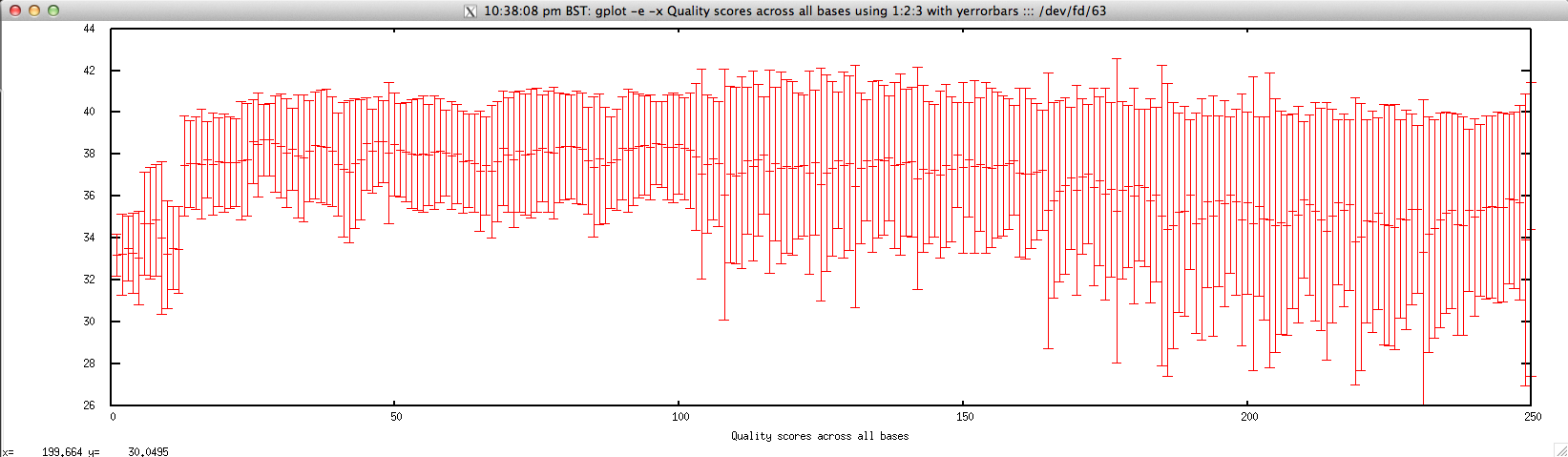
- Per-base quality score for FASTQ file
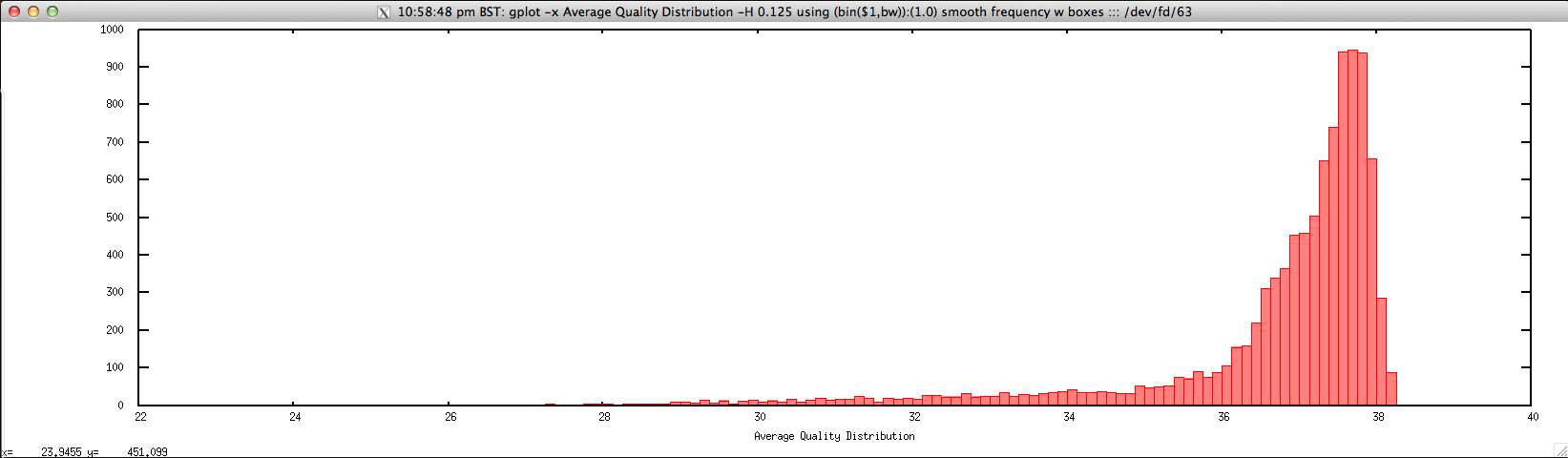
- Average quality distribution for FASTQ file
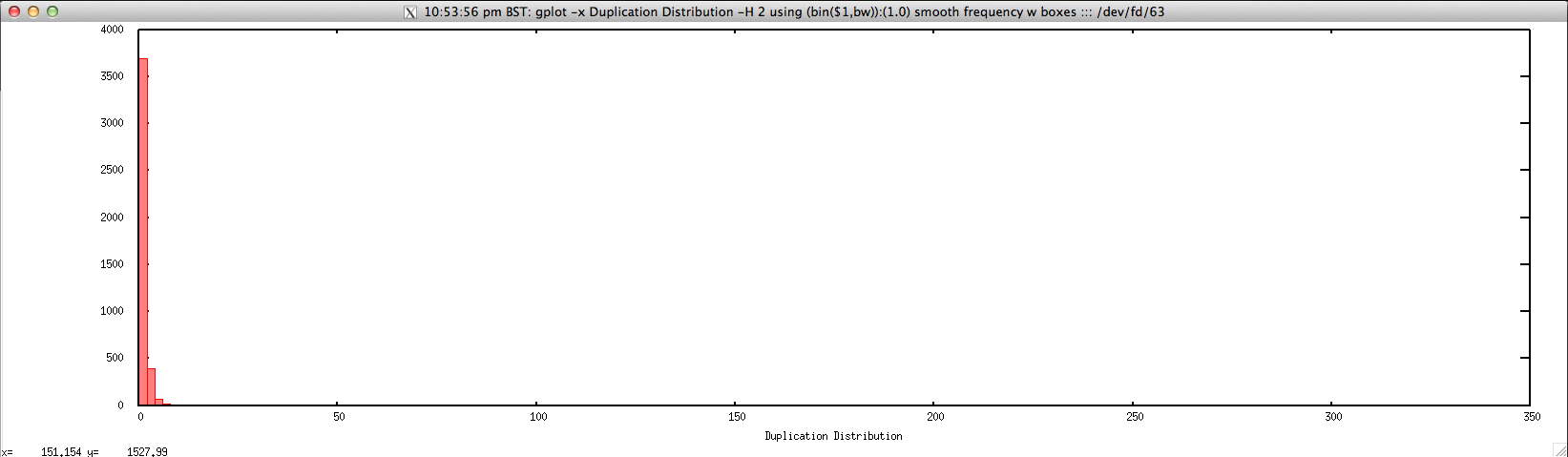
- Duplication distribution for FASTQ/FASTA file
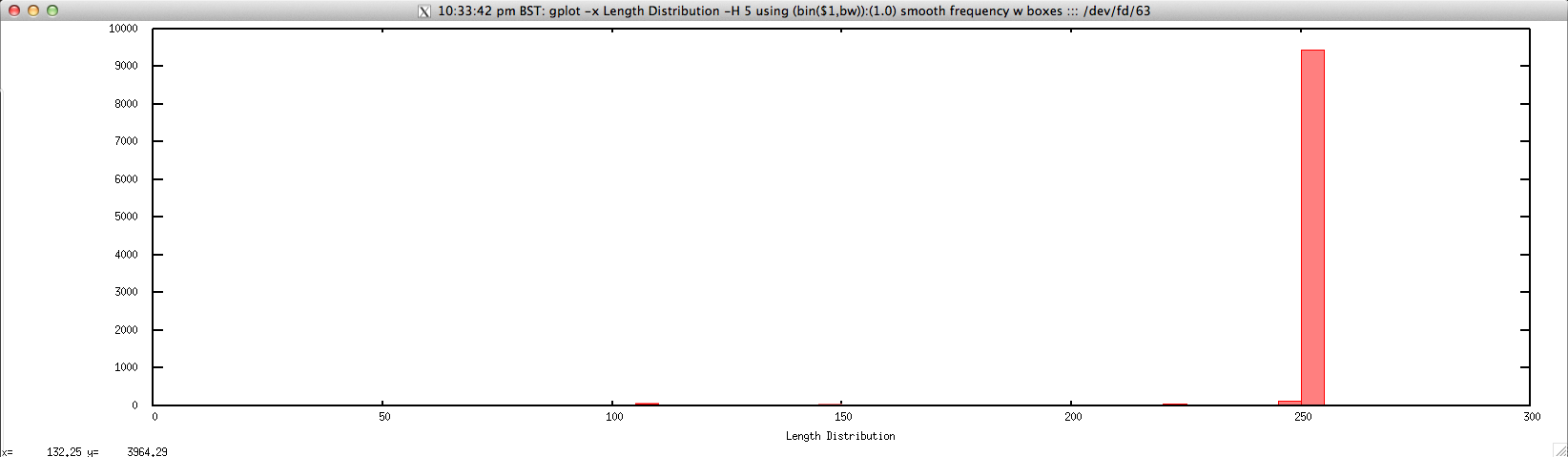
- Length distribution for FASTQ/FASTA file
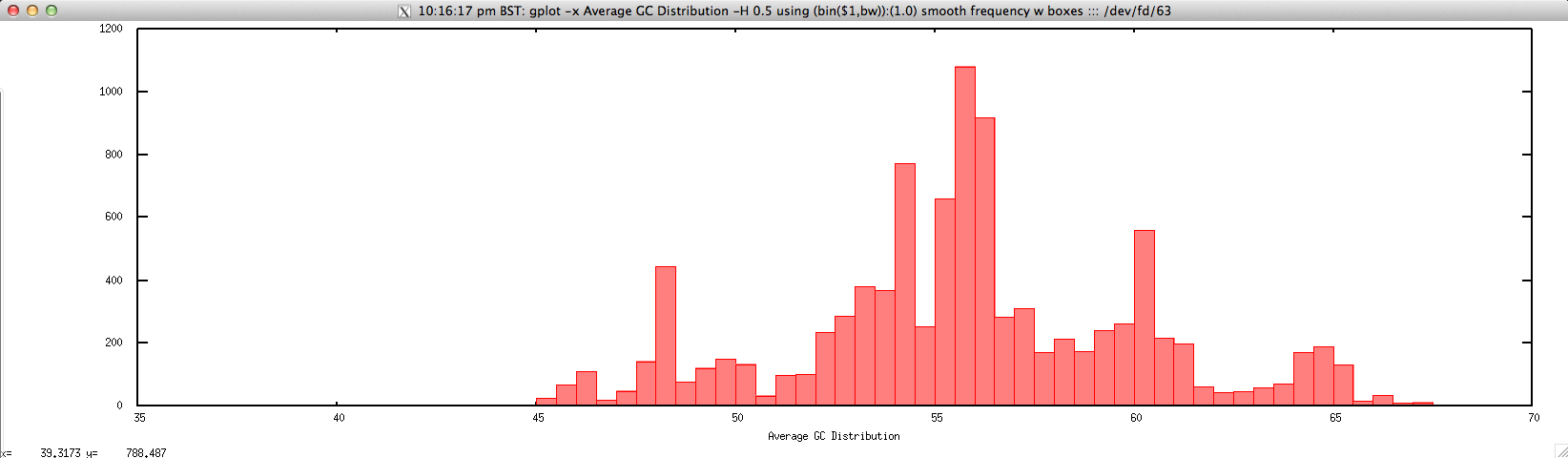
- Average GC distribution for FASTQ/FASTA file
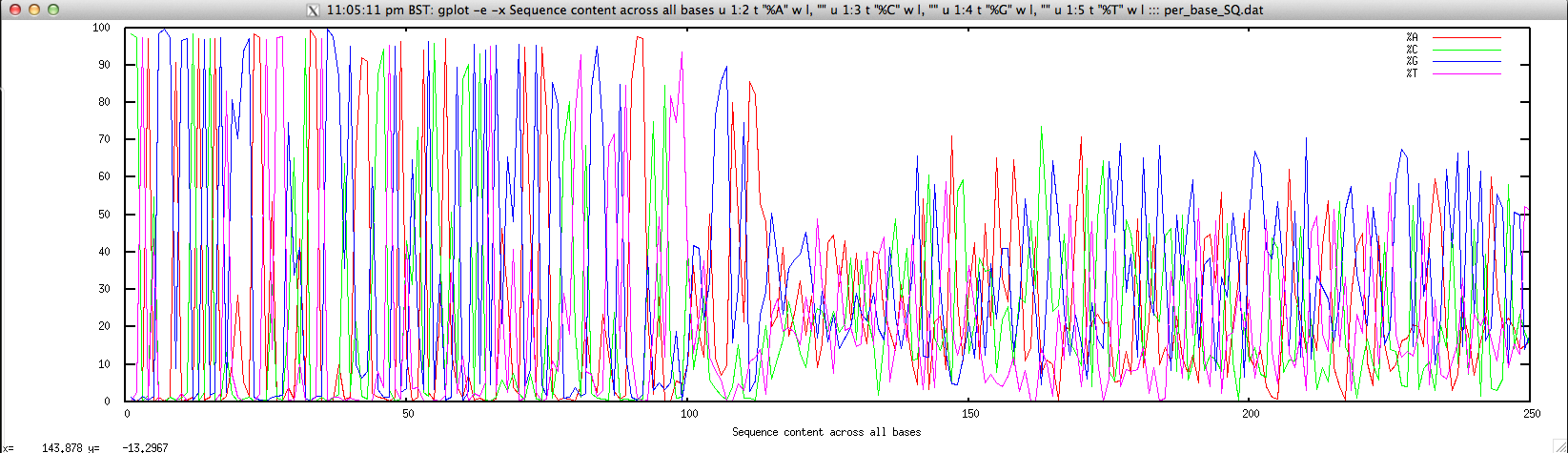
- Per-base sequence content for FASTQ/FASTA file
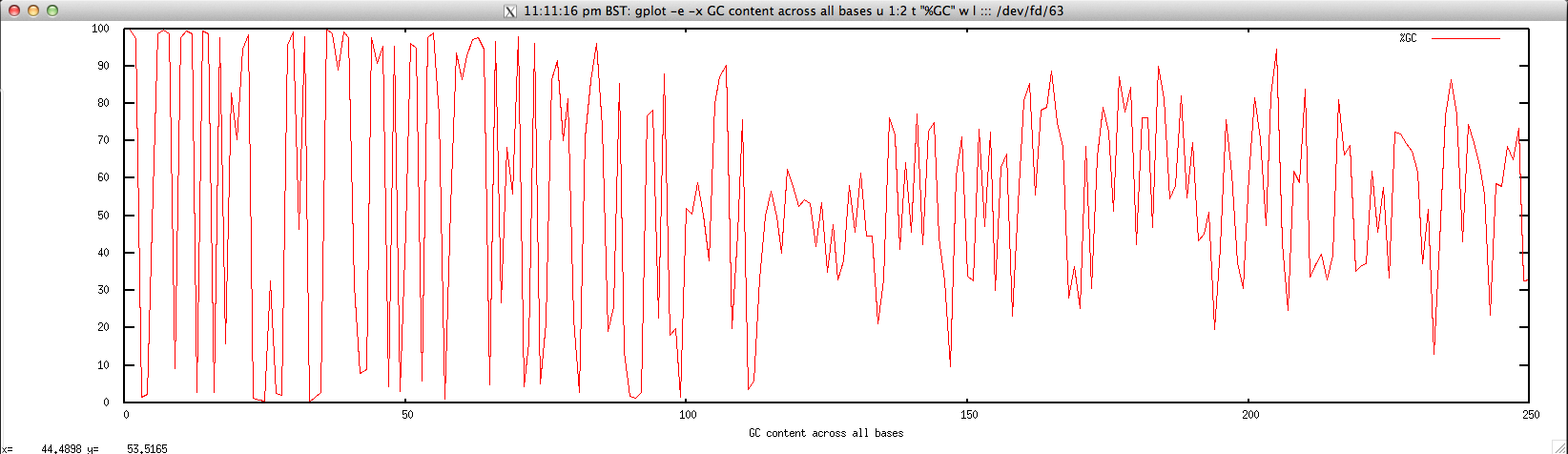
- Per-base GC content for FASTQ/FASTA file
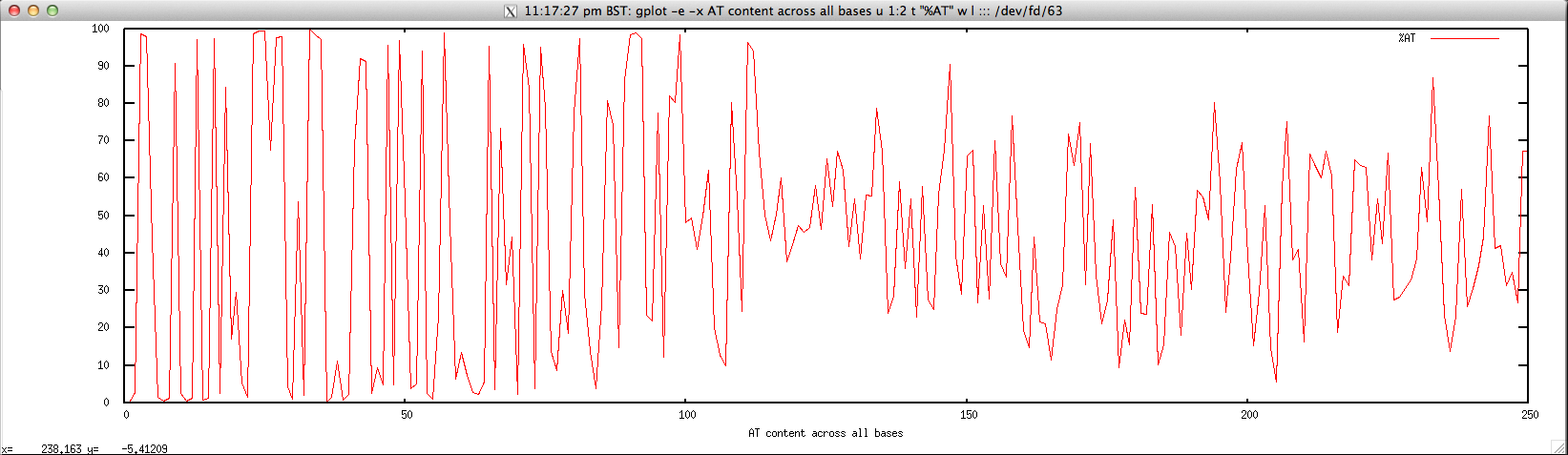
- Per-base AT content for FASTQ/FASTA file
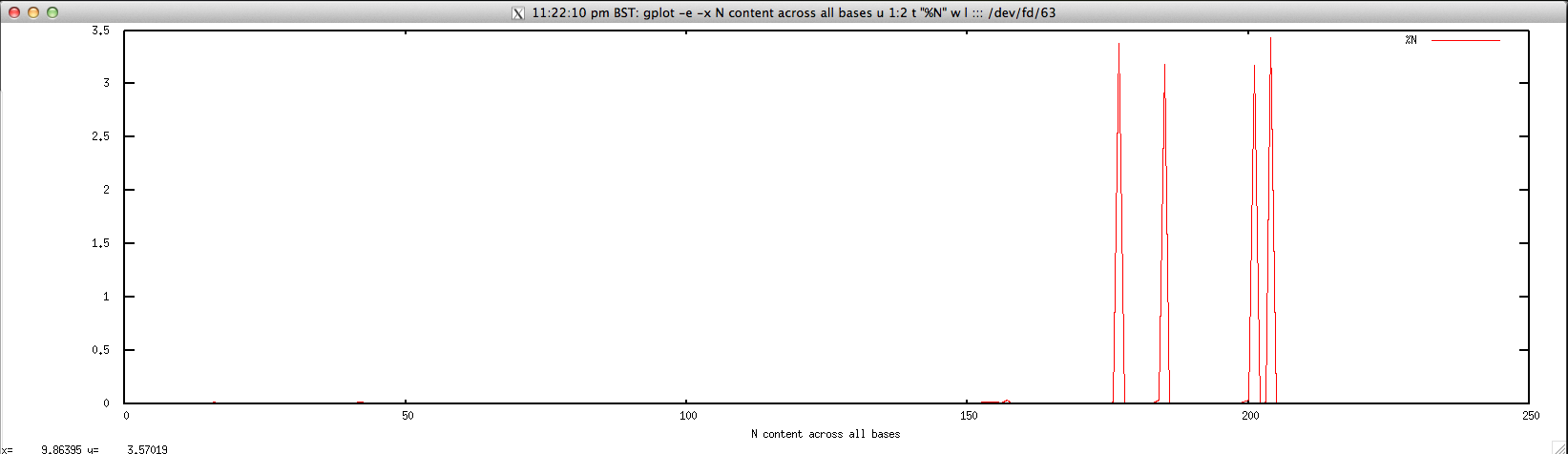
- Per-base N content for FASTQ/FASTA file
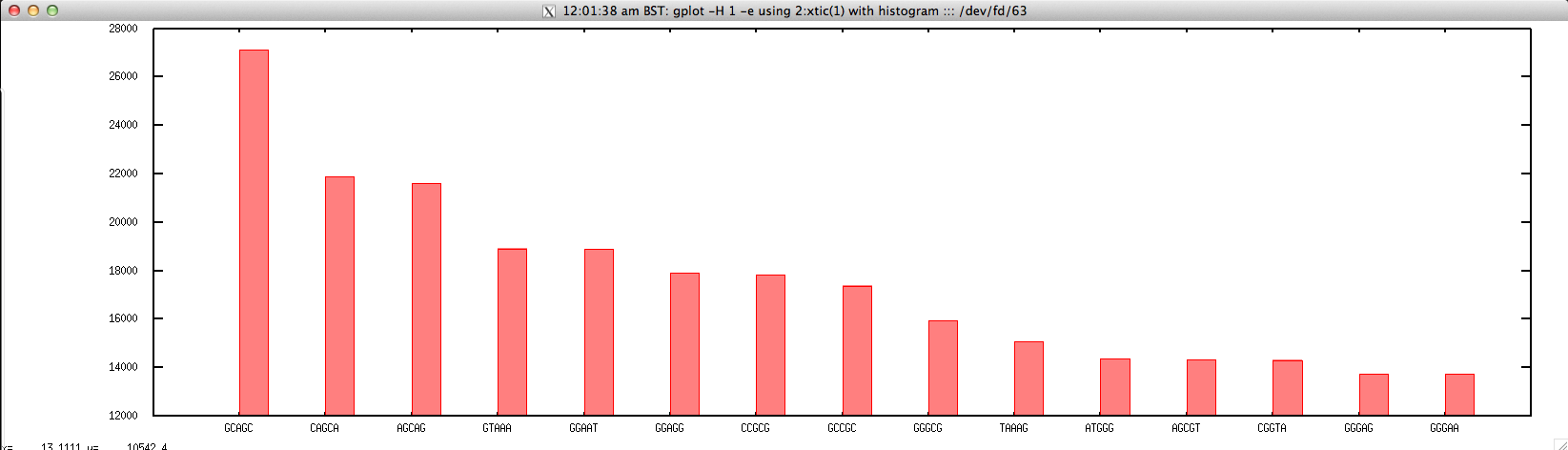
- Top N Kmers for FASTQ/FASTA file
Hopefully, in coming days, I'll update the webpage to explain the underlying logic and be more clear on what I have done.
Best Wishes,
Umer

