Edit (09/20): Also be sure to check out a new promising tool https://github.com/const-ae/glmGamPoi
glmGamPoi estimates dispersion and fits the GLM to single-cell data conceptually similar to DESeq2/edgeR but with very notable gains in speed. It also implements an edgeR-like quasilikelihood ratio test, therefore could be a replacement for edgeR or DESeq2 on single-cell data that scales much better with the number of cells.
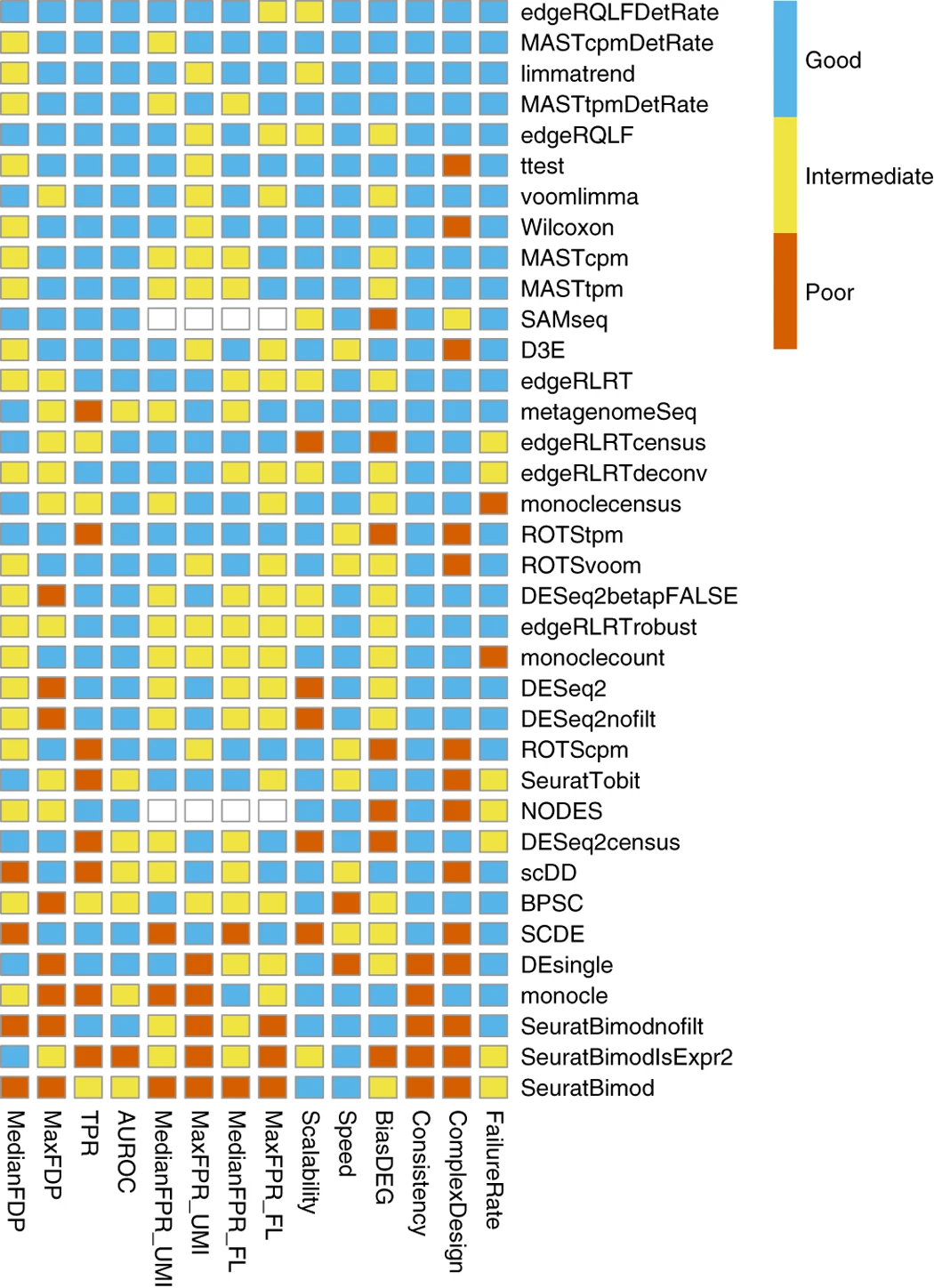
Refreshing this some years later, look at this benchmarking study which includes edgeR, DESeq2 and limma.
https://www.nature.com/articles/nmeth.4612
Code for all tested pipelines is available: https://github.com/csoneson/conquer_comparison/tree/master/scripts
The key take-home messages from this paper for me were:
- methods developed for bulk RNA-seq do not perform worse, often even better than dedicated single-cell methods.
- Wilcox and T-Tests perform well overall but do not allow for complex designs so these are probably limited to simple two-group pairwise comparisons such as marker gene detection within clusters of the same dataset.
- prefiltering of genes with low overall expression is beneficial for some methods such as edgeR. In the linked study the authors removed genes with a TPM below 1 in more than 25% of all cells.
- including the cellular detection rate ("the fraction of detected genes per cell", original quote from the paper) into the design formula is beneficial for some methods such as edgeR
- overall the edgeR QLF pipeline when filtering for lowly-expressed genes and including the cellular detection rate into the design ranks among the best tested methods in this setup
The code for the top-ranked edgeRQLFDetRate is here: https://github.com/csoneson/conquer_comparison/blob/master/scripts/apply_edgeRQLFDetRate.R
Still, I guess it strongly depends on the dataset how each method performs and we are still lacking gold standard benchmarking datasets with wetlab-confirmed true-positive and negative differential genes to robustly benchmark DE
methods.
I personally prefer to aggregate clusters to pseudobulks and then run e.g. edgeR as a pseudobulk comparison, e.g. 2 vs 2 given that you have (here n=2 per group) experimental replicates for your scRNA-seq data. This is (in my hands) much more robust, based on meeting anticipated results and expected pathway enrichment. But this is only based on my dataset so not sure one can generalize.
This table ranks all tested tools and pipelines towards performance in this study with their datasets and their choice of parameters. I strongly suggest to explore things yourself on your data.



Use monocle for single-cell RNA-seq
Thanks dariober for your response.
I'm aware of Monocle and am currently using it. But what I wanted to know is that can the standard RNA-seq tools be used for single-cell RNA-seq?Are there no specific requirements for single-cell rna-seq in terms of normalization etc, given that single-cell has the information of a single transcriptome whereas the standard rna-seq gives the average expression pattern of a cell-population.