I'd like to create an HTML file (from the XML file and XSL stylesheet) similar to what It can be achieved when we performed a BLAST search on the NCBI server. What is the use of this file (xml) is the most appropriate or should i use another kind of out of blast (ASN. ...), and are there any examples of such a XSL file to format .
9
Entering edit mode
14.1 years ago
Pierre Lindenbaum
165k
Update: I wrote the XSLT stylesheet. It is available on github.
Example: The XML:
(..)
<Hit>
<Hit_num>2</Hit_num>
<Hit_id>gi|27881483|ref|NM_017590.4|</Hit_id>
<Hit_def>Homo sapiens zinc finger CCCH-type containing 7B (ZC3H7B), mRNA</Hit_def>
<Hit_accession>NM_017590</Hit_accession>
<Hit_len>5868</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>114.899</Hsp_bit-score>
<Hsp_score>126</Hsp_score>
<Hsp_evalue>1.2149e-23</Hsp_evalue>
<Hsp_query-from>896</Hsp_query-from>
<Hsp_query-to>1302</Hsp_query-to>
<Hsp_hit-from>2927</Hsp_hit-from>
<Hsp_hit-to>2518</Hsp_hit-to>
<Hsp_query-frame>1</Hsp_query-frame>
<Hsp_hit-frame>-1</Hsp_hit-frame>
<Hsp_identity>280</Hsp_identity>
<Hsp_positive>280</Hsp_positive>
<Hsp_gaps>17</Hsp_gaps>
<Hsp_align-len>417</Hsp_align-len>
<Hsp_qseq>CCTTCTGGGCAGG---TGCCATTCATATACCTATCACAAATACTGAAATAGCCTGTTGGGAAGCGGTGCTGCCAGCAGTACTGGTCGTCC---TCGGTGTGGAAAAC
CTTCTCTTTGTGCTTCTCGGAGGAGATGTGGCCCTGCCACTGCTTCTCACTGTTGCAGTTTTTCCCACACATCCAGCAGTGAAAGTCCACTGTAACTTCAGCATAATCTGTTGGCATGTGAAT
TTG-TTTTCCATTTTCCTTGTTTGACTGACTGGCTAT---GTCTTCACTTTTTTCATTTTTTTGACTCTTGAGCCATAGTTCGTAAAATTGCTCCATATCTTGTATCCCATTCTCCTTCATGT
AAGTCCAAACTTCTCTTTCCTCAGGACTATGAGCAAAGGAACAGTTTCCAACATATTGACATTT</Hsp_qseq>
<Hsp_hseq>CCATCTGGGCAGGCTTTGCCCTTCTGGAGCCTGTCGCAGAGCCGGAACTCGCCCATGGGGAAGCGGAAGGCCCAGCCG--CTGG-CGTCACTGTCGGACGTGAAGAC
CTTCTCCTTGTGCTTCTCGGACTGGATGTGCTGCTGCCACTGCTTCTTGCTGTTGCTGTTCTTGCCGCAGAGCCAGCAGTGGTAGCCCATCATGATGTCCGCGTAGTCCGTGGGCATCTGGAT
CTGCTTCTCCCCTTCCCGAG---AACTGA-TGGGGGTCCCTTCTCCAGGCTTTCCTGGGTTGTGTTTTTTCAGCCACATGTCATAGGTCTGCTGCATGTCCAGGATCTTGTTCTCCTTCATGA
AGGTCCACATGTCCCTCTCCTCCGGGCTGTGTGCGAAGGAGCAGTTCCCCACATATTGGCACTT</Hsp_hseq>
<Hsp_midline>|| |||||||||| |||| ||| ||| || || | | ||| | ||| | ||||||||| ||||| | |||| |||| |||| |||
|||||||| |||||||||||||| |||||| |||||||||||||| ||||||| ||| || || || | ||||||||| || ||| | | || || || || || ||||| ||
|| || || ||| || || | ||||| ||| | ||| || ||| | || || | || ||||| | || || |||| ||| || | ||| ||||||||||
|| | ||||| | || || ||||| || || || || ||||| ||||| || |||||||| || ||</Hsp_midline>
</Hsp>
</Hit_hsps>
(...)
Run the stylesheet:
xsltproc --novalid blast2html.xsl blast.xml
Result:
(...)
>gi|27881483|ref|NM_017590.4||NM_017590|Homo sapiens zinc finger CCCH-type containing 7B (ZC3H7B), mRNA
Length=5868
Score = 114.899 bits (126), Expect = 1.2149e-23
Identities = 280/417 (67.1462829736211%), Gaps = 17/417 (4.07673860911271%)
Strand = Plus/Minus
Query 896 CCTTCTGGGCAGG---TGCCATTCATATACCTATCACAAATACTGAAATAGCCTGTTGGG 952
|| |||||||||| |||| ||| ||| || || | | ||| | ||| | |||
Sbjct 2927 CCATCTGGGCAGGCTTTGCCCTTCTGGAGCCTGTCGCAGAGCCGGAACTCGCCCATGGGG 2868
Query 953 AAGCGGTGCTGCCAGCAGTACTGGTCGTCC---TCGGTGTGGAAAACCTTCTCTTTGTGC 1009
|||||| ||||| | |||| |||| |||| ||| |||||||| ||||||
Sbjct 2867 AAGCGGAAGGCCCAGCCG--CTGG-CGTCACTGTCGGACGTGAAGACCTTCTCCTTGTGC 2811
Query 1010 TTCTCGGAGGAGATGTGGCCCTGCCACTGCTTCTCACTGTTGCAGTTTTTCCCACACATC 1069
|||||||| |||||| |||||||||||||| ||||||| ||| || || || | |
Sbjct 2810 TTCTCGGACTGGATGTGCTGCTGCCACTGCTTCTTGCTGTTGCTGTTCTTGCCGCAGAGC 2751
Query 1070 CAGCAGTGAAAGTCCACTGTAACTTCAGCATAATCTGTTGGCATGTGAATTTG-TTTTCC 1128
|||||||| || ||| | | || || || || || ||||| || || || || |||
Sbjct 2750 CAGCAGTGGTAGCCCATCATGATGTCCGCGTAGTCCGTGGGCATCTGGATCTGCTTCTCC 2691
Query 1129 ATTTTCCTTGTTTGACTGACTGGCTAT---GTCTTCACTTTTTTCATTTTTTTGACTCTT 1185
|| || | ||||| ||| | ||| || ||| | || || | ||
Sbjct 2690 CCTTCCCGAG---AACTGA-TGGGGGTCCCTTCTCCAGGCTTTCCTGGGTTGTGTTTTTT 2635
Query 1186 GAGCCATAGTTCGTAAAATTGCTCCATATCTTGTATCCCATTCTCCTTCATGTAAGTCCA 1245
||||| | || || |||| ||| || | ||| |||||||||||| | |||||
Sbjct 2634 CAGCCACATGTCATAGGTCTGCTGCATGTCCAGGATCTTGTTCTCCTTCATGAAGGTCCA 2575
Query 1246 AACTTCTCTTTCCTCAGGACTATGAGCAAAGGAACAGTTTCCAACATATTGACATTT 1302
| || || ||||| || || || || ||||| ||||| || |||||||| || ||
Sbjct 2574 CATGTCCCTCTCCTCCGGGCTGTGTGCGAAGGAGCAGTTCCCCACATATTGGCACTT 2518
(...)
I wrote a stylesheet to display the blast output as SVG+XHTML. Transforming to XHTML should be even easier. See my post.
or a quick search in google returned this page
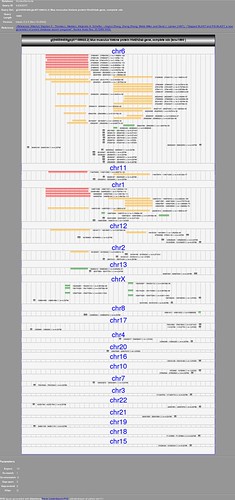
4
Entering edit mode
14.0 years ago
Pablo Pareja
★
1.6k
Hi all,
I implemented some time ago several Java XML wrapper classes for dealing with XML output from Blast.
They are available in this GitHub repository, (this project also includes many other general bioinformatics related XML wrappers).
These are some of the elements modelled:
Hope this can be useful.
Cheers,
Pablo
Similar Posts
Error! readyState=0 status=error text=
Loading Similar Posts
Traffic: 2538 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


why did I get a downvote ? :-)
Many thanks Pierre
Hi Pierre,
This looks like an excellent xsl ... does it still work? I tried running the command you detailed against a blastn xml and it is returning many errors.
Many thanks,
Sean
For example (below). Any pointers would be much appreciated.
Thanks very much, Sean
OK I found a newer version in your Github repo: https://github.com/lindenb/xslt-sandbox
It works a with some issues on Firefox ... Chrome just renders text.
Thanks you.