Start with a cage containing five monkeys.
Inside the cage, hang a banana on a string and place a set of stairs
under it. Before long, a monkey will go to the stairs and start to
climb towards the banana. As soon as he touches the stairs, spray all
of the other monkeys with cold water.
After a while, another monkey makes an attempt with the same result -
all the other monkeys are sprayed with cold water. Pretty soon, when
another monkey tries to climb the stairs, the other monkeys will try
to prevent it.
Now, put away the cold water. Remove one monkey from the cage and
replace it with a new one. The new monkey sees the banana and wants to
climb the stairs. To his surprise and horror, all of the other monkeys
attack him.
After another attempt and attack, he knows that if he tries to climb
the stairs, he will be assaulted.
Next, remove another of the original five monkeys and replace it with
a new one. The newcomer goes to the stairs and is attacked. The
previous newcomer takes part in the punishment with enthusiasm!
Likewise, replace a third original monkey with a new one, then a
fourth, then the fifth. Every time the newest monkey takes to the
stairs, he is attacked.
Most of the monkeys that are beating him have no idea why they were
not permitted to climb the stairs or why they are participating in the
beating of the newest monkey.
After replacing all the original monkeys, none of the remaining
monkeys have ever been sprayed with cold water. Nevertheless, no
monkey ever again approaches the stairs to try for the banana. Why
not? Because as far as they know that's the way it's always been done
round here.
There is no difference between choosing AT ratio and choosing GC ratio, because their sum is 1. So, the choice is merely based on convention.
BTW, the monkey experiment was actually done by a researcher in 1967.

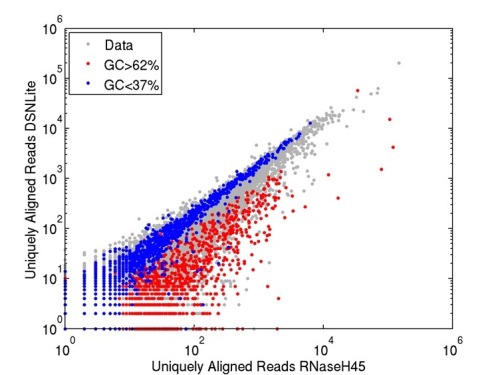

"Why do we normalize for GC counts" : 'normalize' for what ? expression ? ngs ? ...
My bad. Yes, gene expression in RNA-seq. Libraries used to normalize for data (like EDASeq) normalizes for GC content. I'm just curious why GC and not AT.